Training with different topologies#
Int this tutorial, you will learn how to use EncoderMap’s new sparse trainign feature to learn the conformations of multiple proteins with different topologies.
Run this notebook on Google Colab:
Find the documentation of EncoderMap:
https://ag-peter.github.io/encodermap
Install encodermap on Google colab#
If you are on Google colab, please uncomment this line and install encodermap:
[1]:
# !pip install "git+https://github.com/AG-Peter/encodermap.git@main"
# !pip install -r pip install -r https://raw.githubusercontent.com/AG-Peter/encodermap/main/tests/test_requirements.md
Imports#
[2]:
import encodermap as em
import numpy as np
%matplotlib inline
%load_ext autoreload
%autoreload 2
%config Completer.use_jedi = False
2023-02-07 11:08:13.846687: I tensorflow/core/platform/cpu_feature_guard.cc:193] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 AVX512F FMA
To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
2023-02-07 11:08:14.013145: W tensorflow/compiler/xla/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libcudart.so.11.0'; dlerror: libcudart.so.11.0: cannot open shared object file: No such file or directory; LD_LIBRARY_PATH: /opt/hostedtoolcache/Python/3.9.16/x64/lib
2023-02-07 11:08:14.013172: I tensorflow/compiler/xla/stream_executor/cuda/cudart_stub.cc:29] Ignore above cudart dlerror if you do not have a GPU set up on your machine.
2023-02-07 11:08:14.860026: W tensorflow/compiler/xla/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libnvinfer.so.7'; dlerror: libnvinfer.so.7: cannot open shared object file: No such file or directory; LD_LIBRARY_PATH: /opt/hostedtoolcache/Python/3.9.16/x64/lib
2023-02-07 11:08:14.860121: W tensorflow/compiler/xla/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libnvinfer_plugin.so.7'; dlerror: libnvinfer_plugin.so.7: cannot open shared object file: No such file or directory; LD_LIBRARY_PATH: /opt/hostedtoolcache/Python/3.9.16/x64/lib
2023-02-07 11:08:14.860150: W tensorflow/compiler/tf2tensorrt/utils/py_utils.cc:38] TF-TRT Warning: Cannot dlopen some TensorRT libraries. If you would like to use Nvidia GPU with TensorRT, please make sure the missing libraries mentioned above are installed properly.
Fix tensorflow seed for reproducibility
[3]:
import tensorflow as tf
tf.random.set_seed(1)
Load the trajectories#
We use EncoderMap’s TrajEnsemble class to load the trajectories and do the feature alignment.
[4]:
traj_files = ["glu7.xtc", "asp7.xtc"]
top_files = ["glu7.pdb", "asp7.pdb"]
trajs = em.load(traj_files, top_files)
Load the CVs with the ensemble=True options.
[5]:
trajs.load_CVs("all", ensemble=True)
[6]:
trajs
[6]:
<encodermap.TrajEnsemble object. Current backend is no_load. Containing 20002 frames. CV central_distances with shape (20002, 20) loaded. CV central_dihedrals with shape (20002, 18) loaded. CV central_angles with shape (20002, 19) loaded. CV central_cartesians with shape (20002, 21, 3) loaded. CV side_dihedrals with shape (20002, 21) loaded. Object at 0x7f73e4da48b0>
Create the AngleDihedralCartesianEncoderMap#
The AngleDihedralCartesianEncoderMap tries to learn all of the geometric features of a protein. The angles (backbone angles, backbone dihedrals, sidechain dihedrals) are passed through a neuronal network autoencoder, while the distances between the backbone atoms are used to create cartesian coordinates from the learned angles. The generated cartesians and the input (true) cartesians are used to construct pairwise C\(_\alpha\) distances, which are then also weighted using sketchmap’s
sigmoid function. The cartesian_cost_scale_soft_start gradually increases the contribution of this cost function to the overall model loss.
[7]:
p = em.ADCParameters(use_backbone_angles=True,
distance_cost_scale=1,
auto_cost_scale=0.1,
cartesian_cost_scale_soft_start=(50, 80),
n_neurons = [500, 250, 125, 2],
activation_functions = ['', 'tanh', 'tanh', 'tanh', ''],
use_sidechains=True,
summary_step=1,
tensorboard=True,
periodicity=2*np.pi,
n_steps=100,
checkpoint_step=1000,
dist_sig_parameters = (4.5, 12, 6, 1, 2, 6),
main_path=em.misc.run_path('runs/asp7_glu7_asp8'),
model_api='functional',
)
emap = em.AngleDihedralCartesianEncoderMap(trajs, p)
Output files are saved to runs/asp7_glu7_asp8/run0 as defined in 'main_path' in the parameters.
Input contains nans. Using sparse network.
2023-02-07 11:08:20.661900: W tensorflow/compiler/xla/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libcuda.so.1'; dlerror: libcuda.so.1: cannot open shared object file: No such file or directory; LD_LIBRARY_PATH: /opt/hostedtoolcache/Python/3.9.16/x64/lib
2023-02-07 11:08:20.661947: W tensorflow/compiler/xla/stream_executor/cuda/cuda_driver.cc:265] failed call to cuInit: UNKNOWN ERROR (303)
2023-02-07 11:08:20.661972: I tensorflow/compiler/xla/stream_executor/cuda/cuda_diagnostics.cc:156] kernel driver does not appear to be running on this host (fv-az267-630): /proc/driver/nvidia/version does not exist
2023-02-07 11:08:20.662284: I tensorflow/core/platform/cpu_feature_guard.cc:193] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 AVX512F FMA
To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
input shapes are:
{'central_distances': (20002, 20), 'central_dihedrals': (20002, 18), 'central_angles': (20002, 19), 'central_cartesians': (20002, 21, 3), 'side_dihedrals': TensorShape([20002, 21])}
You must install pydot (`pip install pydot`) and install graphviz (see instructions at https://graphviz.gitlab.io/download/) for plot_model to work.
Saved a text-summary of the model and an image in runs/asp7_glu7_asp8/run0, as specified in 'main_path' in the parameters.
train
[8]:
emap.train()
100%|██████████| 100/100 [00:45<00:00, 2.20it/s, Loss after step 100=2.89, Cartesian cost Scale=1]
WARNING:absl:Function `_wrapped_model` contains input name(s) args_0 with unsupported characters which will be renamed to args_0_6 in the SavedModel.
WARNING:absl:Found untraced functions such as _update_step_xla while saving (showing 1 of 1). These functions will not be directly callable after loading.
WARNING:absl:Function `_wrapped_model` contains input name(s) args_0 with unsupported characters which will be renamed to args_0_4 in the SavedModel.
WARNING:absl:Function `_wrapped_model` contains input name(s) Decoder_Input with unsupported characters which will be renamed to decoder_input in the SavedModel.
Plot the result#
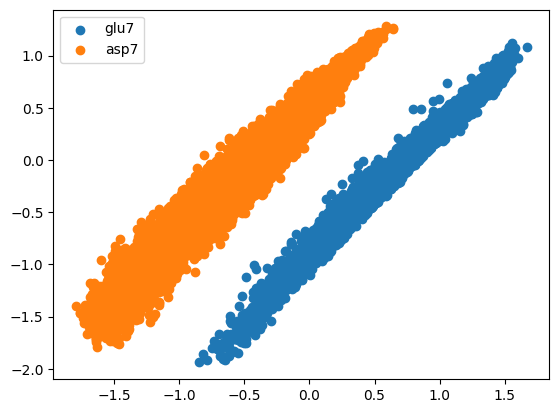
In the result (longer training would be beneficial here), the projection area of asp7 and glu7 are separated.
[9]:
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
# generate ids, based on the names of the trajs
ids = (trajs.name_arr == "asp7").astype(int)
ax.scatter(*emap.encode()[ids == 0].T, label="glu7")
ax.scatter(*emap.encode()[ids == 1].T, label="asp7")
ax.legend()
[9]:
<matplotlib.legend.Legend at 0x7f7352a9a460>
Create a new trajectory#
Using the InteractivePlotting class, we can easily generate new molecular conformations by using the decoder part of the neural network. If you’re running an interactive notebook, you can use the notebook or qt5 backend and play around with the InteractivePlotting.
[10]:
# %matplotlib qt5
%matplotlib inline
sess = em.InteractivePlotting(emap)
Using the `train_data` attribute of `autoencoder`.
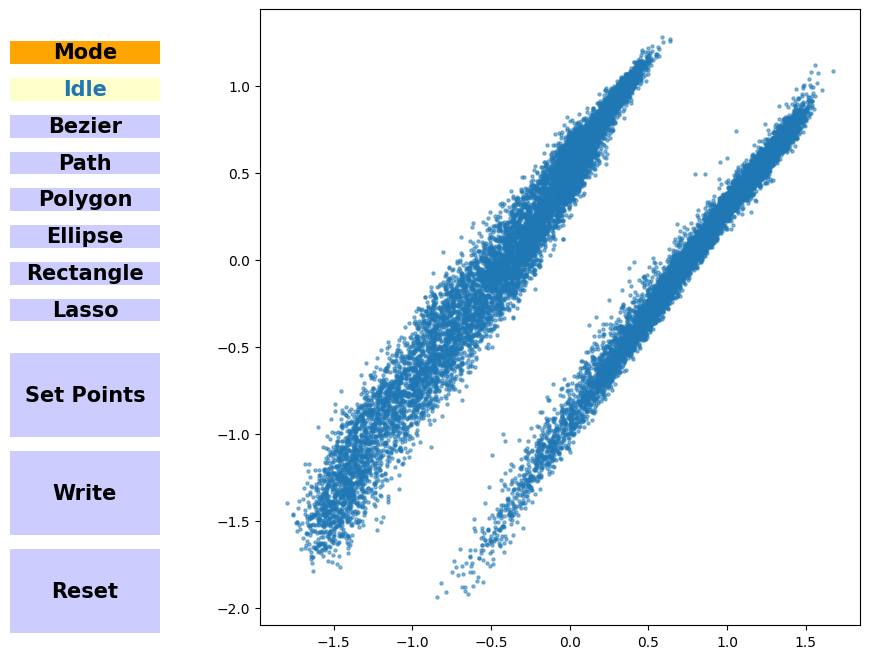
For static notebooks, we load the points along the path and generate new molecular conformations from them.
[11]:
sess.statusmenu.status = "Bezier"
sess.ball_and_stick = True
sess.path_points = np.load("path.npy")
sess.tool.ind = np.load("path.npy")
sess.set_points()
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/MDAnalysis/analysis/base.py:438: UserWarning: Reader has no dt information, set to 1.0 ps
self.times[i] = ts.time
View the generated traj:
[12]:
sess.view
[ ]: