Publication Figure 5#
This jupyter notebook contains the Analysis code for an upcoming publication.
Authors: Kevin Sawade (kevin.sawade@uni-konstanz.de), Tobias Lemke, Christine Peter (christine.peter@uni-konstanz.de)
EncoderMap’s featurization was inspired by the (no longer maintained) PyEMMA library. Consider citing it (https://dx.doi.org/10.1021/acs.jctc.5b00743), if you are using EncoderMap:
Imports#
We start with the imports.
[1]:
# Future imports at the top
from __future__ import annotations
# Import EncoderMap
import encodermap as em
from encodermap.plot import plotting
from encodermap.plot.plotting import _plot_free_energy
# Builtin packages
import re
import io
import warnings
import os
import json
import contextlib
import time
from copy import deepcopy
from types import SimpleNamespace
from pathlib import Path
# Math
import numpy as np
import pandas as pd
import xarray as xr
# ML
import tensorflow as tf
# Plotting
import plotly.express as px
import plotly.graph_objects as go
import plotly.io as pio
from plotly.subplots import make_subplots
import plotly.io as pio
# MD
import mdtraj as md
import MDAnalysis as mda
import nglview as nv
# dates
from dateutil import parser
/home/kevin/git/encoder_map_private/encodermap/__init__.py:194: GPUsAreDisabledWarning: EncoderMap disables the GPU per default because most tensorflow code runs with a higher compatibility when the GPU is disabled. If you want to enable GPUs manually, set the environment variable 'ENCODERMAP_ENABLE_GPU' to 'True' before importing EncoderMap. To do this in python you can run:
import os; os.environ['ENCODERMAP_ENABLE_GPU'] = 'True'
before importing encodermap.
_warnings.warn(
Using Autoreload we can make changes in the EncoderMap source code and use the new code, without needing to restart the Kernel.
[3]:
%load_ext autoreload
%autoreload 2
The autoreload extension is already loaded. To reload it, use:
%reload_ext autoreload
Functions for this notebook.
[4]:
@contextlib.contextmanager
def set_env(**environ):
"""
Temporarily set the process environment variables.
>>> with set_env(PLUGINS_DIR='test/plugins'):
... "PLUGINS_DIR" in os.environ
True
>>> "PLUGINS_DIR" in os.environ
False
:type environ: dict[str, unicode]
:param environ: Environment variables to set
"""
old_environ = dict(os.environ)
os.environ.update(environ)
try:
yield
finally:
os.environ.clear()
os.environ.update(old_environ)
Trained network weights#
EncoderMap’s Neural Network is initialized with random weights and biases. While the training is deterministic with pre-defined training weights, and the inferences that can be made from two trained models are qualitatively similar, the numerical exact output depends on the starting weights.
For this reason, the trained network weights are available from the corresponding authors upon reasonable request or by raising an issue on GitHub: AG-Peter/encodermap#issues
Figure 4 demonstrates how EncoderMap can deal when the training input contains different topologies. The MD simulations used for this figure were orginally conducted for:
@article{kienle2022electrostatic,
title={Electrostatic and steric effects underlie acetylation-induced changes in ubiquitin structure and function},
author={Kienle, Simon Maria and Schneider, Tobias and Stuber, Katrin and Globisch, Christoph and Jansen, Jasmin and Stengel, Florian and Peter, Christine and Marx, Andreas and Kovermann, Michael and Scheffner, Martin},
journal={Nature communications},
volume={13},
number={1},
pages={5435},
year={2022},
publisher={Nature Publishing Group UK London}
}
They can be downloaded from KonDATA as well. First, let use define, where the data will be put.
[5]:
figure_5_data_dir = Path.cwd() / "analysis/figure_5"
if not figure_5_data_dir.exists():
figure_5_data_dir.mkdir(parents=True)
Load MD Data from KonDATA#
[6]:
em.get_from_kondata(
"Ub_K11_mutants",
output=figure_5_data_dir,
silence_overwrite_message=True,
)
[6]:
'/home/kevin/git/encoder_map_private/docs/source/notebooks/publication/analysis/figure_4'
Next, we will load the .xtc and .pdb files into an EncoderMap TrajEnsemble. The list of common strings (common_str) defines, how EncoderMap should group the trajectories.
[7]:
common_str=["Ac", "C", "Q", "R", "wt"]
Then we can use Python pathlib’s glob to search for .xtc and .pdb files.
[8]:
xtc_files = list(figure_5_data_dir.rglob("*.xtc"))
for file in xtc_files:
print(file.relative_to(Path.cwd()))
analysis/figure_4/Ub_K11C_I/traj.xtc
analysis/figure_4/Ub_K11Q_III/traj.xtc
analysis/figure_4/Ub_K11Q_II/traj.xtc
analysis/figure_4/Ub_K11Ac_III/traj.xtc
analysis/figure_4/Ub_wt_II/traj.xtc
analysis/figure_4/Ub_wt_III/traj.xtc
analysis/figure_4/Ub_K11Q_I/traj.xtc
analysis/figure_4/Ub_K11R_I/traj.xtc
analysis/figure_4/Ub_wt_I/traj.xtc
analysis/figure_4/Ub_K11C_II/traj.xtc
analysis/figure_4/Ub_K11R_III/traj.xtc
analysis/figure_4/Ub_K11Ac_II/traj.xtc
analysis/figure_4/Ub_K11C_III/traj.xtc
analysis/figure_4/Ub_K11Ac_I/traj.xtc
analysis/figure_4/Ub_K11R_II/traj.xtc
[9]:
pdb_files = []
for file in xtc_files:
pdb_file = file.with_name("start.pdb")
pdb_files.append(pdb_file)
[10]:
Ub_K11_trajs = em.load(xtc_files, pdb_files, common_str=common_str)
Ub_K11_trajs
[10]:
<encodermap.TrajEnsemble object. Current backend is no_load. Containing 15 trajectories. Common str is ['Ac', 'C', 'Q', 'R', 'wt']. Not containing any CVs. Object at 0x72694ed3bd60>
Add custom topologies#
The one outlier in this dataset is the acetylated lysine. This is a non-standard residue, and thus, we need to tell EncoderMap how to deal with it.
[11]:
custom_aas = {
"KAC": (
"K",
{
"optional_bonds": [
("-C", "N"), # the peptide bond to the previous aa
("N", "CA"),
("N", "H"),
("CA", "C"),
("C", "O"),
("CA", "CB"),
("CB", "CG"),
("CG", "CD"),
("CD", "CE"),
("CE", "NZ"),
("NZ", "HZ"),
("NZ", "CH"),
("CH", "OI2"),
("CH", "CI1"),
("C", "+N"), # the peptide bond to the next aa
("C", "+N"), # the peptide bond to the next aa
],
"CHI1": ["N", "CA", "CB", "CG"],
"CHI2": ["CA", "CB", "CG", "CD"],
"CHI3": ["CB", "CG", "CD", "CE"],
"CHI4": ["CG", "CD", "CE", "NZ"],
"CHI5": ["CD", "CE", "NZ", "CH"],
},
)
}
Ub_K11_trajs.load_custom_topology(custom_aas)
Load CVs#
CVs for this dataset can be loaded via
[13]:
with warnings.catch_warnings():
warnings.simplefilter("ignore")
Ub_K11_trajs.load_CVs("all", ensemble=True)
Ub_K11_trajs._CVs
[13]:
<xarray.Dataset>
Dimensions: (frame_num: 4001, ATOM: 228, COORDS: 3,
CENTRAL_DISTANCES: 227,
CENTRAL_ANGLES: 226,
CENTRAL_DIHEDRALS: 225,
SIDE_DIHEDRALS: 162, ATOM_NO: 4,
traj_num: 15)
Coordinates:
* frame_num (frame_num) int64 0 1 2 ... 3999 4000
* ATOM (ATOM) <U3 '1' '2' '3' ... '227' '228'
* COORDS (COORDS) <U10 'POSITION X' ... 'POSIT...
* CENTRAL_DISTANCES (CENTRAL_DISTANCES) <U19 'CENTERDISTA...
* CENTRAL_ANGLES (CENTRAL_ANGLES) <U19 'CENTERANGLE ...
* CENTRAL_DIHEDRALS (CENTRAL_DIHEDRALS) <U19 'CENTERDIH P...
* SIDE_DIHEDRALS (SIDE_DIHEDRALS) <U18 'SIDECHDIH CHI1...
* ATOM_NO (ATOM_NO) int64 0 1 2 3
* traj_num (traj_num) int64 0 1 2 3 ... 11 12 13 14
traj_name (traj_num) <U4 'traj' 'traj' ... 'traj'
time (traj_num, frame_num) float32 0.0 ......
Data variables:
central_cartesians (traj_num, frame_num, ATOM, COORDS) float32 ...
central_distances (traj_num, frame_num, CENTRAL_DISTANCES) float32 ...
central_angles (traj_num, frame_num, CENTRAL_ANGLES) float32 ...
central_dihedrals (traj_num, frame_num, CENTRAL_DIHEDRALS) float32 ...
side_dihedrals (traj_num, frame_num, SIDE_DIHEDRALS) float32 ...
central_cartesians_feature_indices (traj_num, ATOM) int32 0 4 9 ... 762 763
central_distances_feature_indices (traj_num, CENTRAL_DISTANCES, ATOM_NO) float64 ...
central_angles_feature_indices (traj_num, CENTRAL_ANGLES, ATOM_NO) float64 ...
central_dihedrals_feature_indices (traj_num, CENTRAL_DIHEDRALS, ATOM_NO) int32 ...
side_dihedrals_feature_indices (traj_num, SIDE_DIHEDRALS, ATOM_NO) float64 ...
Attributes:
length_units: nm
time_units: ps
feature_axes: ['ATOM', 'CENTRAL_ANGLES', 'CENTRAL_DIHEDRALS', 'CENTRAL...
angle_units: rad
full_paths: ['/home/kevin/git/encoder_map_private/docs/source/notebo...
topology_files: ['/home/kevin/git/encoder_map_private/docs/source/notebo...Train / Load trained network#
The next step requires us to set the corresponding parameters. A description of the parameters is also available.
[14]:
print(em.ADCParameters().parameters)
Parameter | Value | Description
--------------------------------+--------------------------+---------------------------------------------------
n_neurons | [128, 128, 2] | List containing number of neurons for each layer
| | up to the bottleneck layer. For example [128, 128,
| | 2] stands for an autoencoder with the following
| | architecture {i, 128, 128, 2, 128, 128, i} where i
| | is the number of dimensions of the input data.
| | These are Input/Output Layers that are not
| | trained.
--------------------------------+--------------------------+---------------------------------------------------
activation_functions | ['', 'tanh', 'tanh', ''] | List of activation function names as implemented
| | in TensorFlow. For example: "relu", "tanh",
| | "sigmoid" or "" to use no activation function. The
| | encoder part of the network takes the activation
| | functions from the list starting with the second
| | element. The decoder part of the network takes the
| | activation functions in reversed order starting
| | with the second element form the back. For example
| | ["", "relu", "tanh", ""] would result in a
| | autoencoder with {"relu", "tanh", "", "tanh",
| | "relu", ""} as sequence of activation functions.
--------------------------------+--------------------------+---------------------------------------------------
periodicity | 6.283185307179586 | Defines the distance between periodic walls for
| | the inputs. For example 2pi for angular values in
| | radians. All periodic data processed by EncoderMap
| | must be wrapped to one periodic window. E.g. data
| | with 2pi periodicity may contain values from -pi
| | to pi or from 0 to 2pi. Set the periodicity to
| | float("inf") for non-periodic inputs.
--------------------------------+--------------------------+---------------------------------------------------
learning_rate | 0.001 | Learning rate used by the optimizer.
--------------------------------+--------------------------+---------------------------------------------------
n_steps | 1000 | Number of training steps.
--------------------------------+--------------------------+---------------------------------------------------
batch_size | 256 | Number of training points used in each training
| | step
--------------------------------+--------------------------+---------------------------------------------------
summary_step | 10 | A summary for TensorBoard is writen every
| | summary_step steps.
--------------------------------+--------------------------+---------------------------------------------------
checkpoint_step | 5000 | A checkpoint is writen every checkpoint_step
| | steps.
--------------------------------+--------------------------+---------------------------------------------------
dist_sig_parameters | (4.5, 12, 6, 1, 2, 6) | Parameters for the sigmoid functions applied to
| | the high- and low-dimensional distances in the
| | following order (sig_h, a_h, b_h, sig_l, a_l, b_l)
--------------------------------+--------------------------+---------------------------------------------------
distance_cost_scale | | Adjusts how much the distance based metric is
| | weighted in the cost function.
--------------------------------+--------------------------+---------------------------------------------------
auto_cost_scale | | Adjusts how much the autoencoding cost is weighted
| | in the cost function.
--------------------------------+--------------------------+---------------------------------------------------
auto_cost_variant | mean_abs | defines how the auto cost is calculated. Must be
| | one of: * `mean_square` * `mean_abs` * `mean_norm`
--------------------------------+--------------------------+---------------------------------------------------
center_cost_scale | 0.0001 | Adjusts how much the centering cost is weighted in
| | the cost function.
--------------------------------+--------------------------+---------------------------------------------------
l2_reg_constant | 0.001 | Adjusts how much the L2 regularisation is weighted
| | in the cost function.
--------------------------------+--------------------------+---------------------------------------------------
gpu_memory_fraction | | Specifies the fraction of gpu memory blocked. If
| | set to 0, memory is allocated as needed.
--------------------------------+--------------------------+---------------------------------------------------
analysis_path | | A path that can be used to store analysis
--------------------------------+--------------------------+---------------------------------------------------
id | | Can be any name for the run. Might be useful for
| | example for specific analysis for different data
| | sets.
--------------------------------+--------------------------+---------------------------------------------------
model_api | functional | A string defining the API to be used to build the
| | keras model. Defaults to `sequntial`. Possible
| | strings are: * `functional` will use keras'
| | functional API. * `sequential` will define a keras
| | Model, containing two other models with the
| | Sequential API. These two models are encoder and
| | decoder. * `custom` will create a custom Model
| | where even the layers are custom.
--------------------------------+--------------------------+---------------------------------------------------
loss | emap_cost | A string defining the loss function. Defaults to
| | `emap_cost`. Possible losses are: *
| | `reconstruction_loss` will try to train output ==
| | input * `mse`: Returns a mean squared error loss.
| | * `emap_cost` is the EncoderMap loss function.
| | Depending on the class `Autoencoder`, `Encodermap,
| | `ADCAutoencoder`, different contributions are used
| | for a combined loss. Autoencoder uses atuo_cost,
| | reg_cost, center_cost. EncoderMap class adds
| | sigmoid_loss.
--------------------------------+--------------------------+---------------------------------------------------
training | auto | A string defining what kind of training is
| | performed when autoencoder.train() is callsed. *
| | `auto` does a regular model.compile() and
| | model.fit() procedure. * `custom` uses gradient
| | tape and calculates losses and gradients manually.
--------------------------------+--------------------------+---------------------------------------------------
batched | True | Whether the dataset is batched or not.
--------------------------------+--------------------------+---------------------------------------------------
tensorboard | | Whether to print tensorboard information. Defaults
| | to False.
--------------------------------+--------------------------+---------------------------------------------------
seed | | Fixes the state of all operations using random
| | numbers. Defaults to None.
--------------------------------+--------------------------+---------------------------------------------------
current_training_step | | The current training step. Aids in reloading of
| | models.
--------------------------------+--------------------------+---------------------------------------------------
write_summary | | If True writes a summar.txt of the models into
| | main_path if `tensorboard` is True, summaries will
| | also be written.
--------------------------------+--------------------------+---------------------------------------------------
trainable_dense_to_sparse | | When using different topologies to train the
| | AngleDihedralCartesianEncoderMap, some inputs
| | might be sparse, which means, they have missing
| | values. Creating a dense input is done by first
| | passing these sparse tensors through
| | `tf.keras.layers.Dense` layers. These layers have
| | trainable weights, and if this parameter is True,
| | these weights will be changed by the optimizer.
--------------------------------+--------------------------+---------------------------------------------------
using_hypercube | | This parameter is not meant to be set by the user.
| | It allows us to print better error messages when
| | re-loading and re-training a model. It contains a
| | boolean whether a model has been trained on the
| | hypercube example data. If your data is
| | 4-dimensional and you reload a model and forget to
| | prvide your data, the model will happily train
| | with the hypercube (and not your) data. This
| | variable implements a check.
--------------------------------+--------------------------+---------------------------------------------------
track_clashes | | Whether to track the number of clashes during
| | training. The average number of clashes is the
| | average number of distances in the reconstructed
| | cartesian coordinates with a distance smaller than
| | 1 (nm). Defaults to False.
--------------------------------+--------------------------+---------------------------------------------------
track_RMSD | | Whether to track the RMSD of the input and
| | reconstructed cartesians during training. The
| | RMSDs are computed along the batch by minimizing
| | the .. math:: \text{RMSD}(\mathbf{x},
| | \mathbf{x}^{\text{ref}}) = \min_{\mathsf{R},
| | \mathbf{t}} % \sqrt{\frac{1}{N} \sum_{i=1}^{N}
| | \left[ % (\mathsf{R}\cdot\mathbf{x}_{i}(t) +
| | \mathbf{t}) - \mathbf{x}_{i}^{\text{ref}}
| | \right]^{2}} This results in n RMSD values, where
| | n is the size of the batch. A mean RMSD of this
| | batch and the values for this batch will be logged
| | to tensorboard.
--------------------------------+--------------------------+---------------------------------------------------
cartesian_pwd_start | | Index of the first atom to use for the pairwise
| | distance calculation.
--------------------------------+--------------------------+---------------------------------------------------
cartesian_pwd_stop | | Index of the last atom to use for the pairwise
| | distance calculation.
--------------------------------+--------------------------+---------------------------------------------------
cartesian_pwd_step | | Step for the calculation of paiwise distances.
| | E.g. for a chain of atoms N-C_a-C-N-C_a-C...
| | cartesian_pwd_start=1 and cartesian_pwd_step=3
| | will result in using all C-alpha atoms for the
| | pairwise distance calculation.
--------------------------------+--------------------------+---------------------------------------------------
use_backbone_angles | | Allows to define whether backbone bond angles
| | should be learned (True) or if instead mean values
| | should be used to generate conformations (False).
--------------------------------+--------------------------+---------------------------------------------------
use_sidechains | | Whether sidechain dihedrals should be passed
| | through the autoencoder.
--------------------------------+--------------------------+---------------------------------------------------
angle_cost_scale | | Adjusts how much the angle cost is weighted in the
| | cost function.
--------------------------------+--------------------------+---------------------------------------------------
angle_cost_variant | mean_abs | Defines how the angle cost is calculated. Must be
| | one of: * "mean_square" * "mean_abs" *
| | "mean_norm".
--------------------------------+--------------------------+---------------------------------------------------
angle_cost_reference | 1 | Can be used to normalize the angle cost with the
| | cost of same reference model (dummy).
--------------------------------+--------------------------+---------------------------------------------------
dihedral_cost_scale | 1 | Adjusts how much the dihedral cost is weighted in
| | the cost function.
--------------------------------+--------------------------+---------------------------------------------------
dihedral_cost_variant | mean_abs | Defines how the dihedral cost is calculated. Must
| | be one of: * "mean_square" * "mean_abs" *
| | "mean_norm".
--------------------------------+--------------------------+---------------------------------------------------
dihedral_cost_reference | 1 | Can be used to normalize the dihedral cost with
| | the cost of same reference model (dummy).
--------------------------------+--------------------------+---------------------------------------------------
side_dihedral_cost_scale | 0.5 | Adjusts how much the side dihedral cost is
| | weighted in the cost function.
--------------------------------+--------------------------+---------------------------------------------------
side_dihedral_cost_variant | mean_abs | Defines how the side dihedral cost is calculated.
| | Must be one of: * "mean_square" * "mean_abs" *
| | "mean_norm".
--------------------------------+--------------------------+---------------------------------------------------
side_dihedral_cost_reference | 1 | Can be used to normalize the side dihedral cost
| | with the cost of same reference model (dummy).
--------------------------------+--------------------------+---------------------------------------------------
cartesian_cost_scale | 1 | Adjusts how much the cartesian cost is weighted in
| | the cost function.
--------------------------------+--------------------------+---------------------------------------------------
cartesian_cost_scale_soft_start | (None, None) | Allows to slowly turn on the cartesian cost. Must
| | be a tuple with (start, end) or (None, None) If
| | begin and end are given, cartesian_cost_scale will
| | be increased linearly in the given range.
--------------------------------+--------------------------+---------------------------------------------------
cartesian_cost_variant | mean_abs | Defines how the cartesian cost is calculated. Must
| | be one of: * "mean_square" * "mean_abs" *
| | "mean_norm".
--------------------------------+--------------------------+---------------------------------------------------
cartesian_cost_reference | 1 | Can be used to normalize the cartesian cost with
| | the cost of same reference model (dummy).
--------------------------------+--------------------------+---------------------------------------------------
cartesian_dist_sig_parameters | (4.5, 12, 6, 1, 2, 6) | Parameters for the sigmoid functions applied to
| | the high- and low-dimensional distances in the
| | following order (sig_h, a_h, b_h, sig_l, a_l,
| | b_l).
--------------------------------+--------------------------+---------------------------------------------------
cartesian_distance_cost_scale | 1 | Adjusts how much the cartesian distance cost is
| | weighted in the cost function.
--------------------------------+--------------------------+---------------------------------------------------
multimer_training | | Experimental feature.
--------------------------------+--------------------------+---------------------------------------------------
multimer_topology_classes | | Experimental feature.
--------------------------------+--------------------------+---------------------------------------------------
multimer_connection_bridges | | Experimental feature.
--------------------------------+--------------------------+---------------------------------------------------
multimer_lengths | | Experimental feature.
--------------------------------+--------------------------+---------------------------------------------------
reconstruct_sidechains | | Whether to also reconstruct sidechains.
[15]:
total_steps = 5000
parameters = em.ADCParameters(
n_steps=total_steps,
main_path=em.misc.run_path(figure_5_data_dir / "runs"),
cartesian_cost_scale=1,
cartesian_cost_variant="mean_abs",
cartesian_cost_scale_soft_start=(
int(total_steps / 10 * 9),
int(total_steps / 10 * 9) + total_steps // 50,
),
cartesian_pwd_start=1,
cartesian_pwd_step=3,
dihedral_cost_scale=1,
dihedral_cost_variant="mean_abs",
distance_cost_scale=0,
cartesian_distance_cost_scale=100,
checkpoint_step=max(1, int(total_steps / 10)),
l2_reg_constant=0.001,
center_cost_scale=0,
tensorboard=True,
summary_step=1,
use_sidechains = True,
side_dihedral_cost_scale = 20,
use_backbone_angles = True,
cartesian_dist_sig_parameters = (7, 6, 3, 1, 2, 3),
track_clashes = True,
)
[16]:
Ub_K11_emap = em.AngleDihedralCartesianEncoderMap(
parameters=parameters,
trajs=Ub_K11_trajs,
)
Output files are saved to /home/kevin/git/encoder_map_private/docs/source/notebooks/publication/analysis/figure_4/runs/run7 as defined in 'main_path' in the parameters.
input shapes are:
{'central_cartesians': (60015, 228, 3), 'central_dihedrals': (60015, 225), 'central_distances': (60015, 227), 'side_dihedrals': TensorShape([60015, 162]), 'central_angles': (60015, 226)}
Saved a text-summary of the model and an image in /home/kevin/git/encoder_map_private/docs/source/notebooks/publication/analysis/figure_4/runs/run7, as specified in 'main_path' in the parameters.
[17]:
Ub_K11_emap.add_images_to_tensorboard()
Logging images with [(60015, 226), (60015, 225), TensorShape([60015, 162])]-shaped data every 1 epochs to Tensorboard at /home/kevin/git/encoder_map_private/docs/source/notebooks/publication/analysis/figure_4/runs/run7
[19]:
if input(
"Training this model takes a lot of computational resources. "
"If you juts want to look at the images, we recommend loading "
"a trained model from the provided data. Train y/N?"
) == "y":
history = Ub_K11_emap.train()
json.dump(history, open(emap.p.main_path / "history.json", 'w'))
else:
figure_5_trained_nn_dir = Path.cwd() / "trained_networks/figure_5"
Ub_K11_emap = em.AngleDihedralCartesianEncoderMap.from_checkpoint(
trajs=Ub_K11_trajs,
checkpoint_path=figure_5_trained_nn_dir,
)
Training this model takes a lot of computational resources. If you juts want to look at the images, we recommend loading a trained model from the provided data. Train y/N?
Seems like the parameter file was moved to another directory. Parameter file is updated ...
Output files are saved to /home/kevin/git/encoder_map_private/docs/source/notebooks/publication/trained_networks/figure_5 as defined in 'main_path' in the parameters.
input shapes are:
{'central_cartesians': (60015, 228, 3), 'central_dihedrals': (60015, 225), 'central_distances': (60015, 227), 'side_dihedrals': TensorShape([60015, 162]), 'central_angles': (60015, 226)}
Saved a text-summary of the model and an image in /home/kevin/git/encoder_map_private/docs/source/notebooks/publication/trained_networks/figure_5, as specified in 'main_path' in the parameters.
Project all data#
The AngleDihedralCartesianEncoderMap can now directly encode/project molecular trajectory data.
[20]:
Ub_K11_trajs.load_CVs(
data=Ub_K11_emap.encode(Ub_K11_trajs),
attr_name="lowd",
)
Due to the size of the provided data 904226, I need to chunk it, which takes longer. Sit back, grab a coffee...
Due to the size of the provided data 904226, I need to chunk it, which takes longer. Sit back, grab a coffee...
Due to the size of the provided data 904226, I need to chunk it, which takes longer. Sit back, grab a coffee...
Due to the size of the provided data 904226, I need to chunk it, which takes longer. Sit back, grab a coffee...
Due to the size of the provided data 904226, I need to chunk it, which takes longer. Sit back, grab a coffee...
Due to the size of the provided data 904226, I need to chunk it, which takes longer. Sit back, grab a coffee...
Due to the size of the provided data 904226, I need to chunk it, which takes longer. Sit back, grab a coffee...
Due to the size of the provided data 904226, I need to chunk it, which takes longer. Sit back, grab a coffee...
Due to the size of the provided data 904226, I need to chunk it, which takes longer. Sit back, grab a coffee...
Due to the size of the provided data 904226, I need to chunk it, which takes longer. Sit back, grab a coffee...
Due to the size of the provided data 904226, I need to chunk it, which takes longer. Sit back, grab a coffee...
Due to the size of the provided data 904226, I need to chunk it, which takes longer. Sit back, grab a coffee...
Due to the size of the provided data 904226, I need to chunk it, which takes longer. Sit back, grab a coffee...
Due to the size of the provided data 904226, I need to chunk it, which takes longer. Sit back, grab a coffee...
Due to the size of the provided data 904226, I need to chunk it, which takes longer. Sit back, grab a coffee...
Images of lowd procetions#
The low-dimensional projection can be colorized accordsing to either the mutation.
[22]:
fig = em.plot.plot_trajs_by_parameter(
Ub_K11_trajs,
parameter="common_str",
type="heatmap",
show=False,
nbins=50,
alpha=0.5,
contourtype="contourf",
)
fig.update_layout(
{
"width": 800,
"height": 800,
"template": "none",
"xaxis": {
"showgrid": False,
"showline": False,
"zeroline": False,
"showticklabels": False,
"ticks": "",
},
"yaxis": {
"showgrid": False,
"showline": False,
"zeroline": False,
"showticklabels": False,
"ticks": "",
},
"plot_bgcolor": "rgba(0, 0, 0, 0)",
"paper_bgcolor": 'rgba(0, 0, 0, 0)',
"title": None,
"legend": {
"font": {
"size": 30,
},
},
}
)
# fig.write_image(figure_5_trained_nn_dir / "lowd.svg", scale=2)
# fig.write_image(figure_5_trained_nn_dir / "lowd.png", scale=2)
fig.show()
And using free energy as coloration.
[23]:
from encodermap.plot.plotting import _plot_free_energy
# for the one bundled as project
# basin_indices = [0, 26, 51, 72, 98]
basin_indices = [0, 21, 32, 48, 62, 99]
fig = go.Figure(
data=[
_plot_free_energy(*Ub_K11_trajs.lowd.T, bins=50),
]
)
layout = {
"width": 800,
"height": 800,
"template": "none",
"title": None,
"xaxis": {
"showgrid": False,
"showline": False,
"zeroline": False,
"title": None,
"showticklabels": False,
"ticks": "",
},
"yaxis": {
"showgrid": False,
"showline": False,
"zeroline": False,
"title": None,
"showticklabels": False,
"ticks": "",
},
"plot_bgcolor": "rgba(0, 0, 0, 0)",
"paper_bgcolor": 'rgba(0, 0, 0, 0)',
}
fig.update_layout(**layout)
fig.show()
Cluster and paths#
Clusters and paths can be generated using EncoderMap’s InteractivePlotting. Note, that clusters are extracted from simulated MD conformations, by RMSD-aligning a subset of the selected points. In contrary, paths represent arbitrary points on the low-dimensional projection of the latent space. EncoderMap’s decoder network can generate molecular conformations from these points.
[24]:
sess = em.InteractivePlotting(
trajs=Ub_K11_trajs,
autoencoder=Ub_K11_emap,
)
Cluster#
A cluster has been selected using EncoderMap’s InteractivePlotting. The selected points are also available upon reasonable request.
[25]:
# np.save(figure_5_trained_nn_dir / "cluster_assignment/membership.npy", Ub_K11_trajs._user_selected_points)
Ub_K11_trajs.load_CVs(
np.load(figure_5_trained_nn_dir / "clusters/membership.npy"),
attr_name="cluster_membership",
)
with set_env(ENCODERMAP_SKIP_SCATTER_SIZE_CHECK="True"):
fig = em.plot.plot_trajs_by_parameter(Ub_K11_trajs, "cluster_membership", cbar=False, show=False)
fig.update_layout(**layout)
# fig.write_image(figure_5_trained_nn_dir / "cluster_selection.svg", scale=2)
# fig.write_image(figure_5_trained_nn_dir / "cluster_selection.png", scale=2)
fig.show()
Next, the cluster can be saved to pdb and then rendered using external tools.
[26]:
cluster = Ub_K11_trajs.cluster(0, n_points=10)
for i, (cs, traj) in enumerate(cluster.join(dict_keys="cs").items()):
traj.save_pdb(str(figure_5_trained_nn_dir / f"clusters/trajs_in_cluster_with_{cs}_topology.pdb"))
[27]:
from IPython.display import Image, display
display(Image(figure_5_trained_nn_dir / "clusters/chimeraX_render.png", width=600))
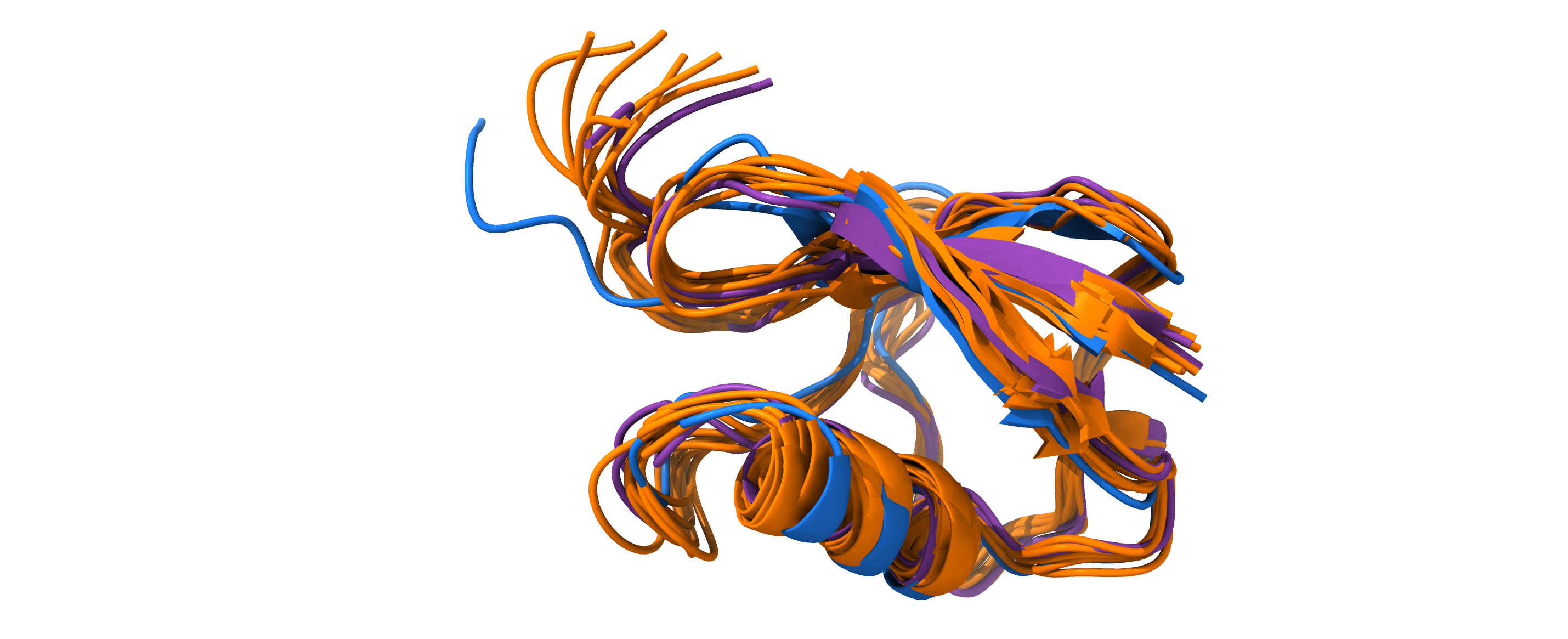
Path#
The selected Path is provided in the accompanzing dataset.
[28]:
# np.save(figure_5_trained_nn_dir / "paths/path.npy", sess.path)
path = np.load(figure_5_trained_nn_dir / "paths/path.npy")
[29]:
from encodermap.plot.plotting import _plot_free_energy
# for the one bundled as project
basin_indices = [0, 21, 32, 48, 62, 99]
fig = go.Figure(
data=[
_plot_free_energy(*Ub_K11_trajs.lowd.T, bins=50),
go.Scatter(
x=path[:, 0],
y=path[:, 1],
mode="lines",
name="",
line_width=5,
customdata=[i for i in range(len(path))],
hovertemplate="%{customdata}",
showlegend=False,
),
go.Scatter(
x=path[basin_indices, 0],
y=path[basin_indices, 1],
mode="markers",
name="",
line_width=0,
showlegend=False,
marker={
"size": 20,
"line": {
"color": "black",
"width": 2,
},
"color": "white",
},
)
]
)
layout = {
"width": 800,
"height": 800,
"template": "none",
"xaxis": {
"showgrid": False,
"showline": False,
"zeroline": False,
"showticklabels": False,
"ticks": "",
},
"yaxis": {
"showgrid": False,
"showline": False,
"zeroline": False,
"showticklabels": False,
"ticks": "",
},
"plot_bgcolor": "rgba(0, 0, 0, 0)",
"paper_bgcolor": 'rgba(0, 0, 0, 0)',
}
fig.update_layout(**layout)
# fig.write_image(figure_5_trained_nn_dir / "path.svg", scale=2)
# fig.write_image(figure_5_trained_nn_dir / "path.png", scale=2)
fig.show()
Generate wt conformations along the path.
[30]:
wt_traj = Ub_K11_emap.generate(path, top="wt")
/home/kevin/git/encoder_map_private/encodermap/trajinfo/trajinfo_utils.py:1299: UserWarning:
Your custom topology for residue name=KAC resSeq=None index=None does not define atoms for the dihedral PSI. If this dihedral consists of standard atom names, it will be considered for dihedral calculations. If this dihedral should not be present in your custom topology you need to explicitly delete it by adding 'PSI_ATOMS': 'delete' to your custom_topology. If you want this dihedral to be present in your topology, you can ignore this warning.
/home/kevin/git/encoder_map_private/encodermap/trajinfo/trajinfo_utils.py:1299: UserWarning:
Your custom topology for residue name=KAC resSeq=None index=None does not define atoms for the dihedral OMEGA. If this dihedral consists of standard atom names, it will be considered for dihedral calculations. If this dihedral should not be present in your custom topology you need to explicitly delete it by adding 'OMEGA_ATOMS': 'delete' to your custom_topology. If you want this dihedral to be present in your topology, you can ignore this warning.
/home/kevin/git/encoder_map_private/encodermap/trajinfo/trajinfo_utils.py:1299: UserWarning:
Your custom topology for residue name=KAC resSeq=None index=None does not define atoms for the dihedral PHI. If this dihedral consists of standard atom names, it will be considered for dihedral calculations. If this dihedral should not be present in your custom topology you need to explicitly delete it by adding 'PHI_ATOMS': 'delete' to your custom_topology. If you want this dihedral to be present in your topology, you can ignore this warning.
[31]:
import ipywidgets as widgets
import nglview as nv
grid = widgets.GridspecLayout(
n_columns=len(basin_indices),
n_rows=1,
justify_content="center",
align_items="center",
height="200px"
)
views = []
for i, b in enumerate(basin_indices):
view = nv.show_mdtraj(wt_traj[b], height="200px", width="200px")
view.clear_representations()
view.add_ribbon(color_scheme="sstruc")
view.add_ball_and_stick(selection="11 and (.CA or .CB. or .CG or .CD or .CE or .NZ)", color="blue")
views.append(view)
grid[0, i] = view
grid
[31]:
Generate different conformations
[32]:
# C
traj_c = Ub_K11_emap.generate(np.expand_dims(path[32], 0), top="C")
print(traj_c.n_atoms)
# wt
traj_wt = Ub_K11_emap.generate(np.expand_dims(path[32], 0), top="wt")
print(traj_wt.n_atoms)
# KAc
traj_KAc = Ub_K11_emap.generate(np.expand_dims(path[32], 0), top="Ac")
print(traj_KAc.n_atoms)
757
762
/home/kevin/git/encoder_map_private/encodermap/trajinfo/trajinfo_utils.py:1299: UserWarning:
Your custom topology for residue name=KAC resSeq=None index=None does not define atoms for the dihedral PSI. If this dihedral consists of standard atom names, it will be considered for dihedral calculations. If this dihedral should not be present in your custom topology you need to explicitly delete it by adding 'PSI_ATOMS': 'delete' to your custom_topology. If you want this dihedral to be present in your topology, you can ignore this warning.
/home/kevin/git/encoder_map_private/encodermap/trajinfo/trajinfo_utils.py:1299: UserWarning:
Your custom topology for residue name=KAC resSeq=None index=None does not define atoms for the dihedral OMEGA. If this dihedral consists of standard atom names, it will be considered for dihedral calculations. If this dihedral should not be present in your custom topology you need to explicitly delete it by adding 'OMEGA_ATOMS': 'delete' to your custom_topology. If you want this dihedral to be present in your topology, you can ignore this warning.
/home/kevin/git/encoder_map_private/encodermap/trajinfo/trajinfo_utils.py:1299: UserWarning:
Your custom topology for residue name=KAC resSeq=None index=None does not define atoms for the dihedral PHI. If this dihedral consists of standard atom names, it will be considered for dihedral calculations. If this dihedral should not be present in your custom topology you need to explicitly delete it by adding 'PHI_ATOMS': 'delete' to your custom_topology. If you want this dihedral to be present in your topology, you can ignore this warning.
763
[33]:
import ipywidgets as widgets
import nglview as nv
grid = widgets.GridspecLayout(
n_columns=3,
n_rows=1,
justify_content="center",
align_items="center",
height="200px"
)
views = []
for i, b in enumerate([traj_c, traj_wt, traj_KAc]):
view = nv.show_mdtraj(b, height="200px", width="200px")
view.clear_representations()
view.add_ribbon(color_scheme="sstruc")
view.add_ball_and_stick(selection="11 and (.CA or .CB. or .CG or .CD or .CE or .NZ)", color="blue")
views.append(view)
grid[0, i] = view
grid
[33]:
[ ]:
